Our research is dedicated to the investigation of the structure-function relationships in RNA, which we pursue by developing novel cross-disciplinary approaches based on mass spectrometry (MS). The greatest biomedical discoveries have always been propelled by major technological advances. At the same time, the greatest technological advances have always been motivated by the need to solve the most challenging biomedical problems. With the overarching goal of supporting the development of new therapeutic strategies, our research focuses on viral systems responsible for infectious diseases and cancer. Current projects involve the investigation of non-protein coding regions of the genomes of SARS-CoV-2, HIV-1, hepatitis C, and Zika virus, which carry out essential regulatory activities in viral replication. Determining the structure/dynamics of these RNAs and their complexes with nucleic acids, proteins, and small molecule ligands can lead to the elucidation of their mechanism of action and the identification of new therapeutic targets. The analysis of RNA post-transcriptional modifications (rPTMs) can provide valuable insights into their regulatory functions and afford the basis for the rapid diagnosis of viral infection. Our projects combine biophysical, computational, bioinformatics, genomics, molecular biology, and virology approaches with new MS techniques to enable the acquisition of otherwise unattainable information and provide the unique insights necessary to move the field forward.
3D Structure Determination
Sample purity, material availability, size limitations, and poor crystallization properties are just some of the factors that may prevent the direct application of classic structural approaches based on NMR and crystallography. We have been developing strategies based on chemical/biochemical probing and high-resolution mass spectrometric detection (i.e., MS3D) for the structural elucidation of samples that are not amenable to classic methods.
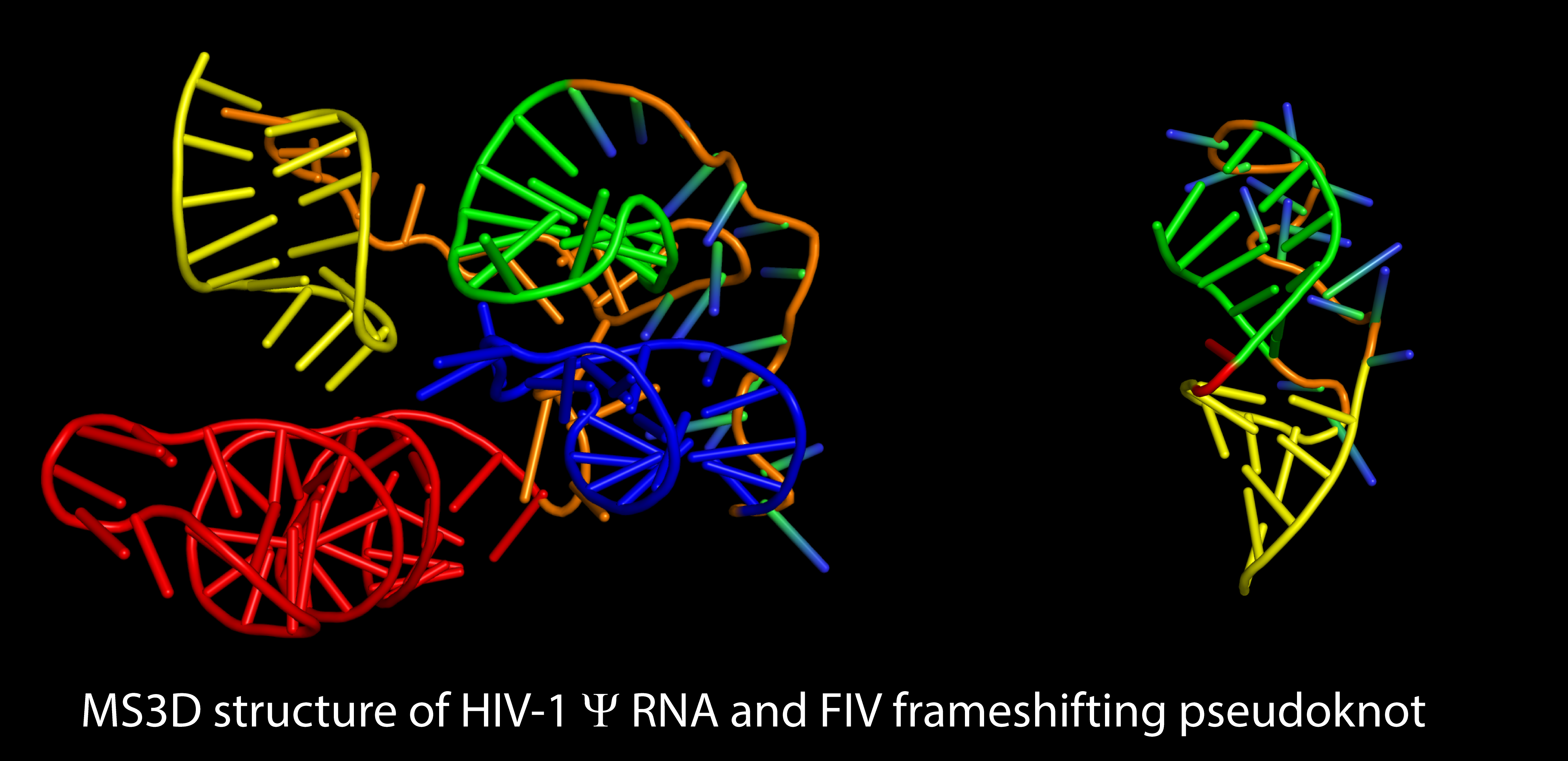
We have employed new crosslinking reagents to obtain valid spatial constraints necessary to support molecular modeling. We have explored structure-specific nucleases that are sensitive to the higher-order structure of RNA substrates. These technologies allowed us to obtain the first full-fledged 3D structures of RNA, which were solved by using experimental constraints obtained by MS-based techniques. We are currently investigating the structure of the frameshifting region of the genome of SARS-CoV-2, which we have found capable of folding into distinctive alternative forms. Comparing such structures could provide the information necessary to design new inhibitors capable of preventing their interconversion.
Conformational Dynamics
The function of non-coding sequences of the human genome, which are estimated to encompass over 98% of the total 3-billion base pairs, is sustained by their ability to fold into well-defined structures capable of interacting with very specific cellular components. We are developing MS-based approaches for investigating the dynamics of long-range interactions that stabilize the higher-order structure of RNA and ribonucleoprotein. Tandem MS allowed us to investigate the process of dimerization and isomerization of the HIV-1 dimerization initiation site (DIS),which is mediated by the viral chaperone nucleocapsid protein (NC).
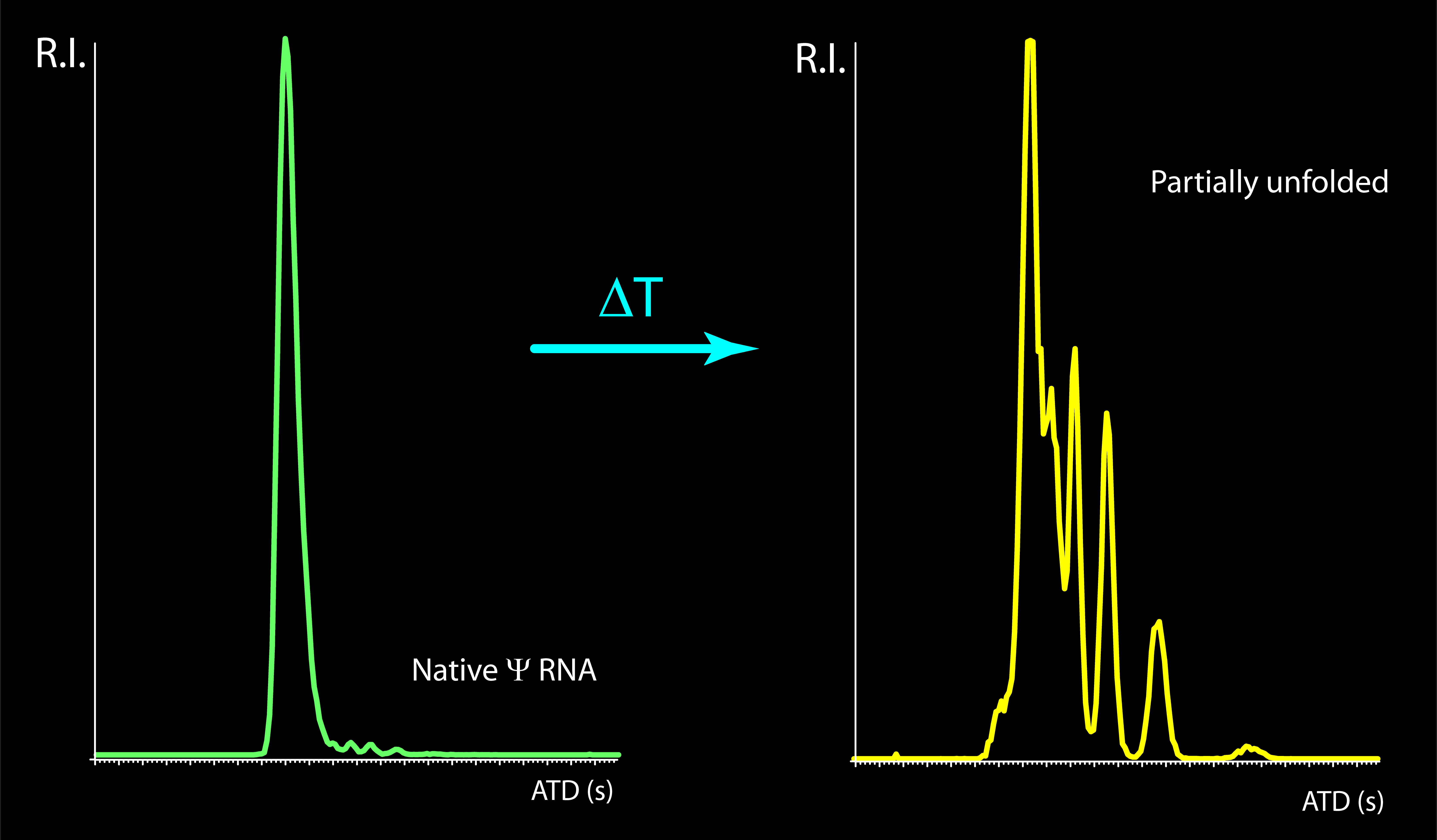
We are now utilizing ion mobility spectrometry (IMS) mass spectrometry to observe the effects of environmental conditions, metals, and other ligands on the stability of RNA structures. We designed a heated-emitter apparatus to observe the unfolding dynamics and determine the conformational stability of RNA. Unlike classic structural methods, IMS-MS enables the simultaneous detection of multiple conformations that are present in solution at the same time. This feature offers the ability to monitor the conformation of interconverting systems, such as riboswitches and ribozymes, which can assume alternative forms during their activity cycle.
Sequencing of mRNAs and viral genomes
In recent years, large, heavily-modified RNAs have been attracting increasing attention by the general public and the pharmaceutical world due to the success of mRNA vaccines in containing the SARS-Cov-2 pandemic. At the same time, the dire need for innovative antiviral therapies capable of circumventing drug resistance and waning vaccine effectiveness has further increased the need to understand the processes involved in the viral life cycle and the emerging roles of RNA post-transcriptional modifications (rPTMs) in mediating essential viral-host interactions (i.e., viral epitranscriptomics). The dearth of genomics approaches for rPTM detection based on next generation sequencing (NGS) affords an excellent opportunity for MS-based approaches, which are capable of differentiating the over 170 natural rPTMs on the basis of unique mass and fragmentation signatures. Our work currently focuses on investigating strategies for increasing the yield of hydrolysis products in the 50-100 nt range, which are ideal for minimizing assignment ambiguities and improving the attainable sequence coverage. The complex mixtures generated by RNA digestions are typically processed by liquid chromatography, while other methods have been overlooked. We are exploring alternative separation techniques, such as capillary zone electrophoresis (CZE), to support the MS analysis of digestion mixtures of increasing complexity obtained from the larger substrates.
Employing specifically designed, highly-selective RNA cleaving deoxyribozymes to reduce the incidence of cleavage events and generate larger digestion products proved effective in fully sequencing a 784 nt-long mRNA without the need for any front-end separation step. On the other hand, limiting the digestion efficiency of more common site-specific RNases and monitoring the reaction process over time by direct infusion MS lead to excellent results in sequencing the same mRNA construct and a large ribosomal RNA (1,869 nt), which presented several rPTMs.
Inhibition Determinants
The binding of proteins, metal ions, and a wide range of ligands can have profound effects on the activity of non-coding RNAs. Evaluating the stability of binding interactions and elucidating their structural determinants are crucial to understand their biological significance. We have developed MS-based approaches for the determination of dissociation constants of protein-RNA and ligand-RNA complexes, which were validated by using isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and fluorescence anisotropy techniques. We have explored the implementation of competitive binding experiments to rank the binding activity of close analogs that vied simultaneously for the same target, thus obtaining valuable information about their binding determinants.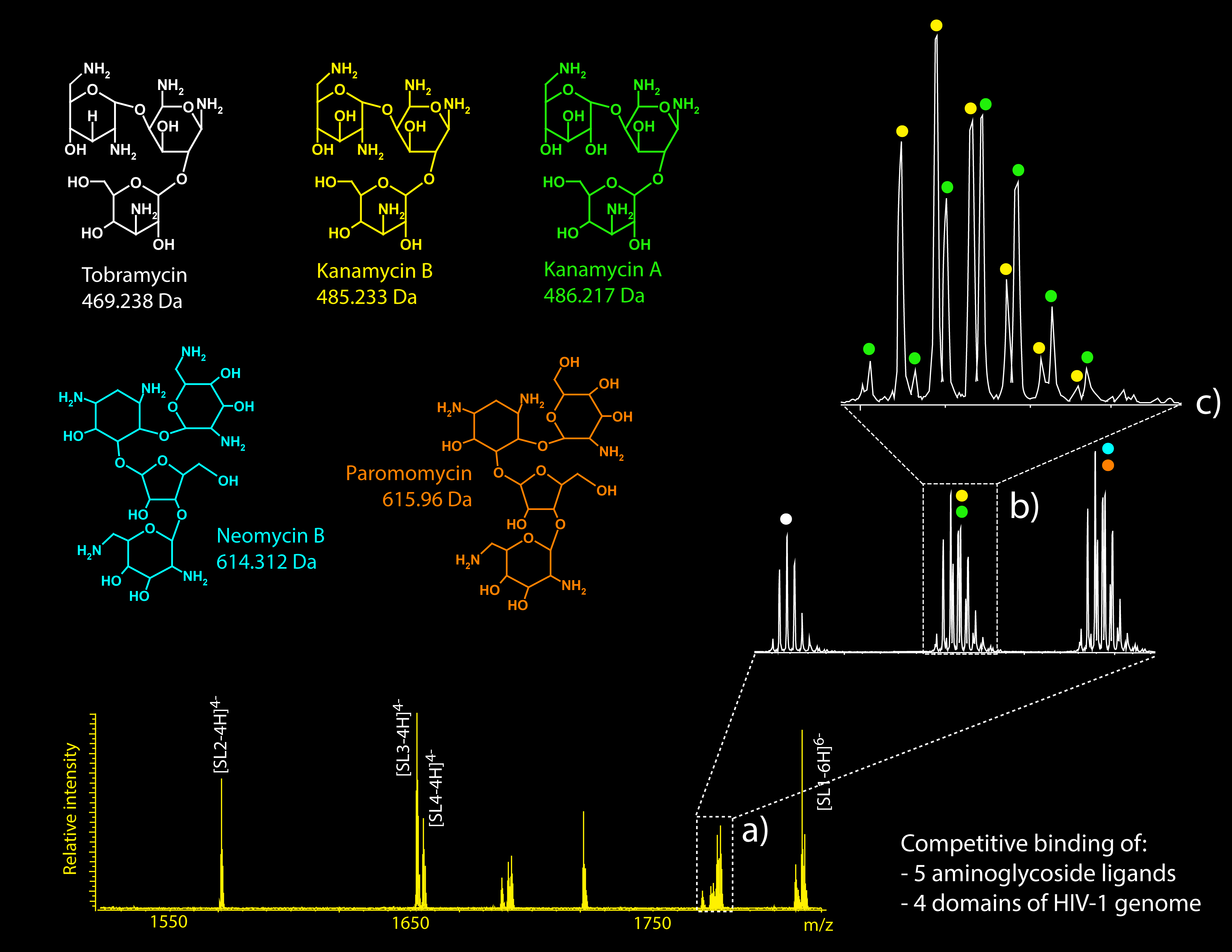
We have developed approaches based on tandem MS, which enable the characterization of binding sites and interfaces between bound components. IMS-MS analyses have shown that the alternative conformers folded by RNA constructs in solution afford remarkably different affinities towards the various ligands, which implies the formation of distinct binding sites with unique characteristics. Additionally, this technique has been providing us with valuable new information on the mechanism of action of nucleic acid chaperones, such as the above-mentioned HIV-1 NC protein and the N-terminal RNA binding domain (NRBD) of the SARS-CoV-2 N-protein.
RNA post-transcriptional modifications and epitranscriptomics regulation
Natural RNA is decorated by over 170 different types of RNA post-transcriptional modifications (rPTMs), which have the ability to affect folding and intermolecular interactions. The fact that such modifications are established, removed, and interpreted by specific protein factors affords the opportunity to place RNA function under the control of signaling pathways involved in the response to stress, infection, and disease. We have developed a novel approach for the global analysis of all the rPTMs expressed by a certain cell at any given time, which allows the investigation of the epitranscriptomics regulation of gene expression. We are employing this approach to explore the possible relationships between essential processes of viral replication and the signaling pathways of different drugs of abuse, which are mediated by a subset of selected rPTMs. This information promises to shed new light onto the mechanism by which such drugs promote the re-emergence of HIV-1 from latency, which could lead to new therapeutic strategies. With collaborators, we are also capitalizing on this approach to understand how hepatitis C and Zika virus employ rPTMs to disguise their genomes and mRNAs as host RNA to elude the innate immune response and take over the normal metabolic machinery.
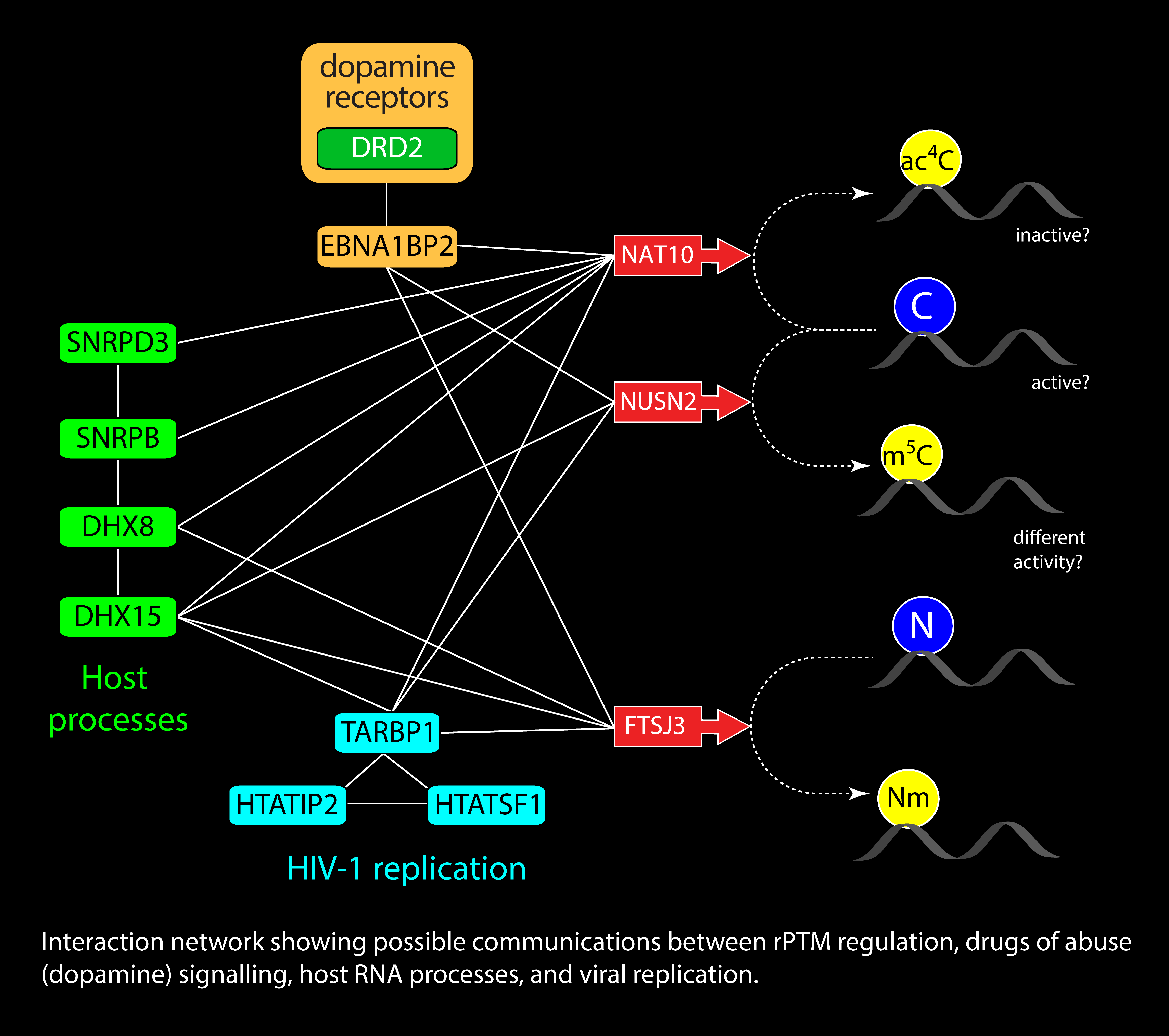
Pathogen Diagnostics
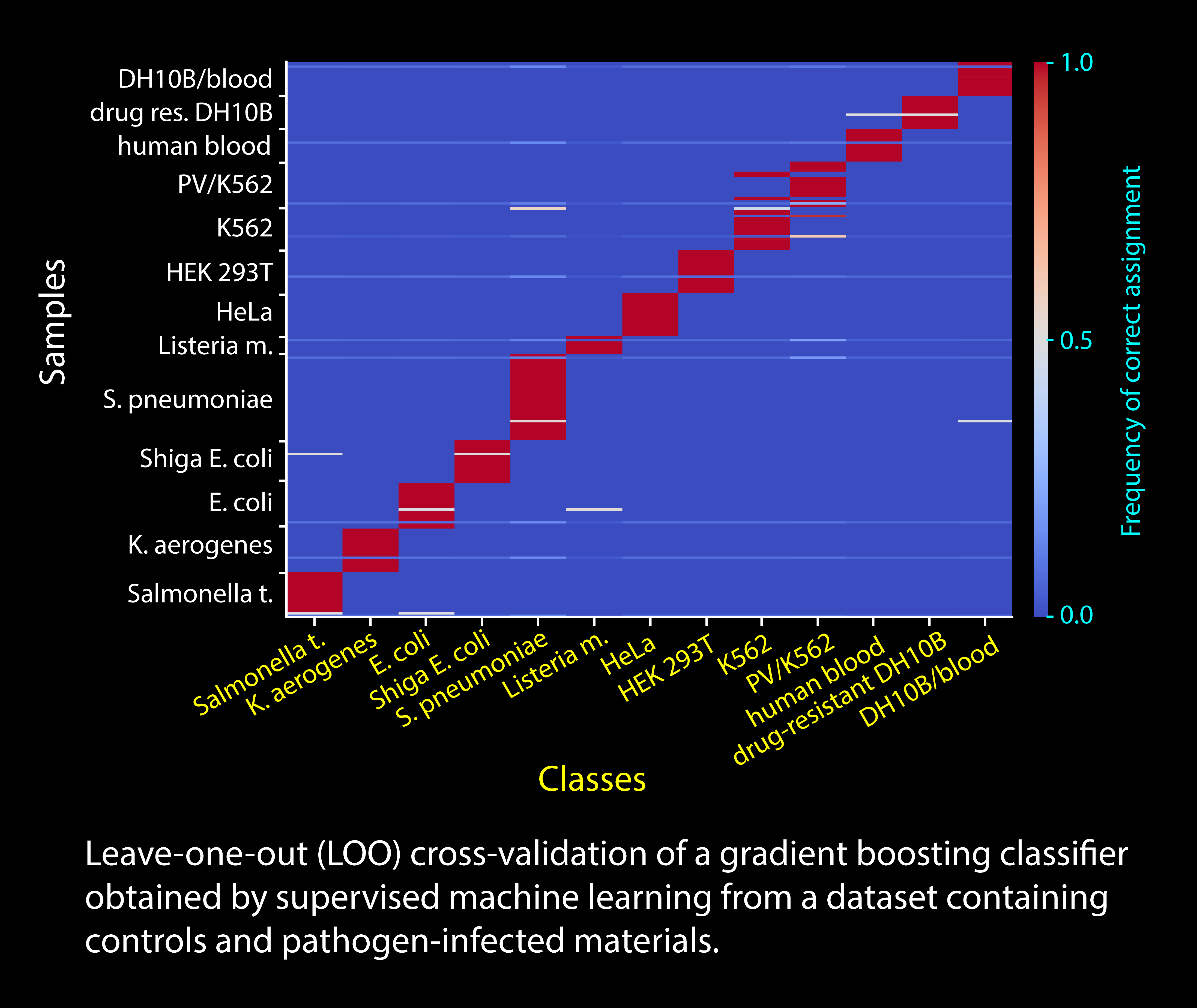
The type and abundance of rPTMs is determined by a combination of factors, including the genetic makeup of the organism under consideration, the possible exposure to environmental factors, and the general metabolic and physiologic state of the cell. This observation affords the opportunity to utilize rPTM profiles obtained from total RNA extracts to evaluate the health of the cell and recognize diseases caused by possible RNA disfunction or viral/bacterial infection. We have developed an approach that combines global rPTM analysis with machine learning (ML) to generate classifiers capable of detecting pathogens in cell cultures and biological fluids. We are now testing the ability of this approach to correctly discriminate samples containing regular or drug-resistant strains. We have recently initiated a collaboration aimed at detecting the causative agents of sepsis in human synovial fluid, which could provide the means to achieve the rapid diagnosis of post-surgery infections necessary to significantly improve patient outcomes. This pilot project will provide proof-of-principle for a wide range of diagnostic applications that would substantiate the definitive transfer of our technology from lab bench to bedside.